Functional Macromolecules Group
About the research group
The Functional Macromolecules and Porous Materials (FunMaP) group specialises on design, synthesis and engineering inclusion compounds, metal complexes, metal-organic frameworks (MOFs), metal organic cages (MOCs) and their derivatives. Our primary focus is on developing eco-friendly and robust materials for applications such as gas capture, separation, catalysis and sensing. We explore material properties starting from understanding their behaviour including molecular interactions, functionality and reactivity. This foundation allows us to tailor materials with desired properties, evaluate the performance for targeted applications and ultimately facilitate implementation in real-world scenarios.
Our team compose with synthetic chemists and engineers both in organic and inorganic areas. We are experienced in a range of synthetic methodologies and perform comprehensive characterization and analysis of material properties using various techniques. In particular at introduction of desired functional components, determination of the molecular and framework structures, study of stability under different conditions, analysis of the optical responses, measurement of accessible surface area, visualisation of macro-scale morphology. We also have hands-on skills to transform materials into practical phase. Our research spans from fundamental studies to industrial implementation. By integrating synthetic principles and programmable functionalities with innovative engineering approaches, the FunMaP group aims to explore new ways to improve material performance and overcome the challenge of existing technologies. Our multidisciplinary research not only enhances the properties of material but also paves the way for the development of sustainable solutions.
Our group also value “QREDIT”: quality, respect, ethics, diversity, innovation and teamwork. With QREDIT, we provide space for researches and personal development. Together, we create a dynamic and inclusive atmosphere where everyone is valued to deliver credible and impactful science.
What are porous materials?
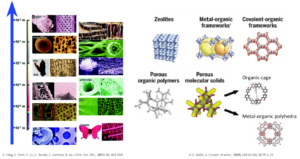
Porous materials usually contain large numbers of voids or pores within their structure/network that allow more efficient mass transfer thanks to their relatively large accessible surface area compared to non-porous materials. Common examples from nature is wood or zeolites. Owing to the specific porous features, porous materials have been extensively studies in decades and have been used in many industrial sectors. Porous materials includes: porous carbon, zeolites, metal organic frameworks (MOFs), covalent organic frameworks (COFs) and cages/polyhedral. Our focus mainly around metal-organic based porous materials. If you are interested, some of these up-to-date journals will provide you more insights.
MOFs are porous infinite structures that composed of metal nodes (i.e. paddle wheel and cluster) and organic linkers (i.e. carboxylate and N-donor ligands) via coordination bonds. Owing to the specific coordination number of metal ions and the geometry of organic linkers. MOFs can be designed following the reticular chemistry and can be functionalised for desired applications.
MOCs (or MOPs) is a discrete cage like supramolecular structures. Similar to MOFs, it is constructed by metal ions and organic linkers but resulting in finite architectures.
Porous liquid (PL) is a new class of materials that was invented in 2007. It has permanent intrinsic porosity in the liquid phase and classified into 3 types. Type 1 PL is the direct liquification of porous solid into liquid via post-functionalisation. Type 2 PL involves the dissolution of porous solid into liquid media and Type 3 PL is the dispersion of porous solid into liquid media. Until recently, a Type 4 PL has been developed, which is the melting of porous solid without losing all the porosity.
Porous gel is a continuous microscopic structure with macroscopic dimensions that is unchanged over a period of time and possesses rheologically solid-like behaviour despite being liquid.
What are macromolecules?
Macromolecules typically composed of numerous of monomers such as polymers or proteins. In biology, they play crucial roles in biological processes. In industry, they impact our daily life. Appropriate design and functionalisation allow implementation of macromolecules in a board range of applications.
Scientists group

Dr. Min Ying Tsang, principal investigator
Head of Functional Macromolecules Research Group
I am currently a Junior Principal investigator of the Functional Macromolecules and Porous Materials Group at the at the Łukasiewicz Research Network – PORT Polish Center for Technology Development. I have extensive experience in design, synthesis and application of various materials. Throughout my career, I focused on development of porous materials including metal-organic frameworks (MOFs), metal organic cages, porous liquids, and lanthanide-based materials and investigate the application potential including water splitting photocatalysis, carbon capture, optical sensing, sensing and germicidal study.
From studying the fundamental phenomenon, I am interested to seek for solutions to create functional porous materials. My international research experience in different countries (Hong Kong, Spain, UK, Japan, Austria, Poland) and participation in national and international conferences also earned me scientific networks from different countries and contacts in cross-disciplinary teams for potential international collaborations and in joint funding acquisitions.
My experience in industrial research and complementation with Master of Business Administration (MBA) degree has also earned me solid experience in technology transfer, commercialisation and outreach to commercial sector in efforts to establish collaborations with industrial partners. Currently, I hold two US patents and more than 20 top-ranked journals. I am also a member of EUROMOF international committee and EU4MOFs COST action. As the leader of the research team, I aim to integrate the multidisciplinary research background of each team members with my vision on implementation of porous materials to tackle current challenges in environmental issues and advance technology development.
ORCID: Min Ying Tsang (0000-0003-4859-5739) – ORCID
LinkedIn: Ivy Tsang – Junior Principal Investigator – Łukasiewicz – PORT | LinkedIn
Google Scholar: Min Ying Tsang profile

Dr. Tapendu Samanta
Postdoctoral scientist
As a synthetic organic chemist specializing in polymer chemistry, my research centers on designing innovative monomers and polymerization techniques to explore and tailor the physicochemical properties of resulting materials for impactful applications. Initially, my work focused on creating fluorogenic and chromogenic polymeric materials as advanced sensing platforms for detecting environmental pollutants. Building on this foundation, I shifted towards sequence-defined polymer synthesis, employing both conventional synthetic methods and automated synthesizers to achieve precision and control.
My current research emphasizes novel synthesis strategies for sequence-defined stereo-controlled oligomers, allowing for fine-tuning of properties for specific uses. Additionally, I am exploring the synthesis of ligands for metal-organic frameworks (MOFs) with adaptable luminescent characteristics. Looking ahead, my goal is to develop polymeric materials with transformative potential in biomedical fields, particularly for bioimaging, drug delivery, and cancer theranostics, ultimately aiming to bridge fundamental research with real-world applications in healthcare and environmental solutions. The purpose of my scientific activity is to advance the synthesis and application of functional materials.
ORCID: https://orcid.org/0000-0002-8542-275X
Google Scholar: https://scholar.google.com/citations?user=O7vxOhIAAAAJ&hl=en&oi=ao
ResearchGate: https://www.researchgate.net/profile/Tapendu-Samanta

Pawel Cywnar
PhD student
I was graduated from Wroclaw University of Science and Technology and from the French university Ecole Normale Superieure de Paris-Saclay. I studied materials science at the Wroclaw University of Science and Technology and Nanobiophotonics in Paris as a part of the Erasmus Mundus Master Programme. My work in PORT has started from September 2020 as a process engineer and later I have started a PhD in the field of sensors for bioactive impurities of water.
Currently, I am working on the development of material for the detection of biologically active water contaminants such as the Bisphenol A(BPA), that is a commonly known endocrine disrupting chemical(EDC). Earlier, I was advancing the one-pot synthesis to facilitate fabrication of sequence-defined oligomers and polymers. Owing to previous projects I have got hands-on experience in organic chemistry, chromatography and mass spectrometry, while right now I learn more skill in optical spectroscopic field.
ORCID: https://orcid.org/0000-0002-7355-9273
 Our group started in 2024 and we are recently at the establishing stage. Currently, we are developing numerous research and industrial projects for national and international fundings. We aim to develop portable sensors to address the current environmental concern. Our current research aims to fabricate materials to effectively detect, capture and eliminate pollutants from air and water. We also investigate highly sensitive and active optical materials such as lanthanide-based metal organic frameworks as optical sensors to detect toxic air pollutants including formaldehyde and VOCs. The high surface area and versatile composition of MOFs allow to be tailored for specific analytes, combine with the predictable lanthanide optical emission properties.
Our group started in 2024 and we are recently at the establishing stage. Currently, we are developing numerous research and industrial projects for national and international fundings. We aim to develop portable sensors to address the current environmental concern. Our current research aims to fabricate materials to effectively detect, capture and eliminate pollutants from air and water. We also investigate highly sensitive and active optical materials such as lanthanide-based metal organic frameworks as optical sensors to detect toxic air pollutants including formaldehyde and VOCs. The high surface area and versatile composition of MOFs allow to be tailored for specific analytes, combine with the predictable lanthanide optical emission properties.
We are experienced in a range of synthetic methodologies including Schlenk technique, extreme temperature (-70 to 300oC) synthesis, air sensitive methods, solvothermal and high pressure autoclave reaction, and mechanochemical synthesis. We also have hand-on skills to perform comprehensive characterization and analysis of material properties using techniques such as nuclear magnetic resonance spectroscopy (NMR), thermogravimetric analysis (TGA), powder x-ray diffractometer (PXRD), chromatography (i.e. HPLC, GC), surface area analysis, scanning electron microscopy (SEM), transmission electron microscopy (TEM), dynamic light scattering (DLS), ultraviolet-visible spectroscopy (UV-Vis), Fluorometer and circular dichroism (CD) spectroscopy.

Specific interests:
- Synthesis and functionalisation of macromolecular complexes and metal-organic frameworks (MOFs) for sensing and photocatalytic applications
- Design new type of metal-organic polyhedral/cages (MOPs/MOCs)
- Develop porous soft materials including porous liquids and porous gels for functional applications such as separation and delivery systems
- Engineer porous materials for industrial applications
Current facilities in FunMaP Lab:

We have state-of-art laboratory and comprehensive facilities to synthesize, characterize porous materials and demonstrate performance, both in solution and solid state.
For synthesis: ovens, Schlenk lines, ball mills
For characterization: UV-Vis spectroscopy, fluorometer, circular dichroism spectroscopy, Flash Differential Scanning Calorimeter (DSC), chromatography, BET surface area analyzer
IP and publications
Macromolecules and Polymers:
Scientific publication highlights:
- N. Das, T. Samanta, S. Rajwar, R. Shunmugam, Biomacromolecules 2024, 25, 2, 990–996.
- N. Das, T. Samanta, D. Patra, P. Kumar, R. Shunmugam, Macromolecules 2024, 57, 3, 976–984.
- P. Cwynar, P. Pasikowski, R. Szweda, Eur. Polym. J. 2023, 182, 111706
- P. Obstarczyk, M. Lipok, A. Żak, P. Cwynar, J. Olesiak-Bańska, Biomater. Sci. 2022, 10(6), 1554-1561
- T. Samanta, N. Das, D. Patra, P. Kumar, B. Sharmistha, R. Shunmugam, ACS Sustainable Chem. Eng. 2021, 9, 14, 5196–5203.
- T. Samanta, N. Das, R. Shunmugam, ACS Sustainable Chem. Eng. 2021, 9, 30, 10176–10183.
- F. Di Salvo, M. Y. Tsang, F. Teixidor, C. Viñas, J. G. Planas, J. Crassous, N. Vanthuyne, N. A. Alcalde, E. Ruiz, G.
- Coquerel, S. Clevers, V. Dupray, D. C. Lazarte, M. E. Light, M. B. Hursthouse, Chem. Eur. J., 2014, 20, 1081.
- M. Y. Tsang, C. Viñas, F. Teixidor, J. G. Planas, N. Conde, R. SanMartin, M. T. Herrero, E. Domínguez, A. Lledós, P. Vidossich, D. Choquesillo-Lazarte, Inorg. Chem., 2014, 53(17), 9284
Patent:
- R. Szweda, P. Walencik, P. Cwynar, M. Turski, „A method of producing polycarbamates with a defined monomeric sequence”, patent application, 2021 PCT/IB2021/057872.
Porous materials:
Scientific publication highlights:
- P.C. Han, C.H. Chuang, S. W. Lin, X. Xiang, Z. Wang, M. Kuzumoto, S. Tokuda, T. Tateishi, A. Legrand, M. Y. Tsang, H.C. Yang, K.C.W. Wu, K. Urayama, D.Y. Kang, S. Furukawa, Nat Commun., 2024, 15, 9523.
- M. Y. Tsang, A. Sinelshchikova, O. Zaremba, F. Schofbeck, A. D. Balsa, M. R. Reithofer, S. Wuttke, J. M. Chin, Adv. Funct. Mater. 2024, 34, 2308376
- M. Y. Tsang, S. Tokuda, P. C. Han, Z. Wang, A. Legrand, M. Kawano, M. Tsujimoto, Y. Ikeno, K. Urayama, K. C.-W. Wu, S. Furukawa, Small Structures, 2022, 3, 2100197.
- M. Y. Tsang, J. P. Convery, B. Lai, J. Cahir, Y. Erbay, D. Rooney, B.Murrer, S. L. James, Materials Today, 2022, 60, 1369.
- E. Sanchez-Gonzalez, M. Y. Tsang, J. Troyano, G. A. Craig, S. Furukawa, Chem. Soc. Rev., 2022, 51, 4876.
- M. Y. Tsang, P. Fałat, M.A. Antoniak, R. Ziniuk, S. J. Zelewski, M. Samoć, M. Nyk, T. Y. Ohulchanskyy, D. Wawrzyńczyk, Nanoscale, 2022, 14, 14770.
- B. Lai, J. Cahir, M. Y. Tsang, J. Jacquemin, D. Rooney, B. Murrer, S. L. James, ACS Appl. Mater. Interfaces, 2021, 13, 932
- J. Cahir, M. Y. Tsang, B.B. Lai, D. Hughes, A. Alam, J. Jacquemin, D. Rooney, S. L. James, Chem. Sci., 2020, 11, 2077
- S. R-Hermida, M. Y. Tsang, C. Vignatti, K. C. Stylianou, V. Guillerm, J. P. Carvajal, F. Teixidor, C. Viñas, D. Choquesillo-Lazarte, C. V. Escamilla, I. Peral, J. Juanhuix, I. Imaz, D. Maspoch, J. G. Planas, Angew. Chem. Int. Ed., 2016, 55, 16049.
- M. Y. Tsang, N. E. Pridmore, L. J. Gillie, Y. H. Chou, R. Brydson, R.E. Douthwaite, Adv. Mater., 2012, 24, 3406
Patent:
- M. Y. Tsang, J. Cahir, D. Rooney, S. L. James, 2018, Porous Liquids, US11565212B2, filed May 2018. Granted January 2023.
- M. Y. Tsang, J. Cahir, S. L. James, 2018, Type-3 Porous Liquids, US11565212B2, filed November 2018. Granted February 2023.
- Starting package (2024-2027): Initialise proof-of-concept findings using porous materials for environmental remediation including sensing device fabrication and visible light driven photocatalytic elimination of pollutants.
- Japan Society for the Promotion of Science (JSPS) fellowship (2019-2020): Development of porous gels and liquids for separation application. The aim was to develop hierarchical porous structure in soft porous materials.
- Co-author of Proof-of-concept grant from Invest Northern Ireland (2016-2019): Development of type-3 porous liquids for effective carbon capture and biogas upgrading. The aim was to formulate simple and cost-effective solution for industrial implementation.
We are open to short-term projects for master students, PhD candidates and postdoctoral researchers to join our research team and explore the innovative potential of porous materials.
There are also schemes to support postdoctoral candidates coming to Poland, if you are interested to join our group, please reach out. Link below is the related funding scheme.
https://nawa.gov.pl/naukowcy/program-im-ulama
Vacancy will also be posted soon!
We are also keen to collaborate with industrial partners to solve current challenges with our materials and technologies.

